Source Nodes
Provide a source of variants
All Variants

All variants in the database.
Cohort

A collection of related samples, eg “control group” or “poor responders”
Classifications

Pedigree
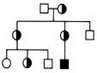
Variants from family samples filtered by genotype according to inheritance models
Sample

A sample, usually one genotype (patient, cell or organism) with a set of variants.
Trio
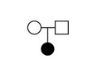
Mother/Father/Proband - filter for recessive/dominant/denovo inheritance
Filter Nodes
These nodes filter variants connected to the top of them
Built In Filter

Built in filters used in node counts eg High or Moderate Impact / OMIM / ClinVar Pathological
Damage

Filter to damage predictions
Filter
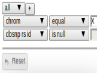
Filter based on column values
Gene List

Filter to a list of gene symbols
Intervals Intersection
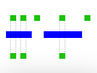
Filter based on intersection with genomic ranges (eg .bed files)
Merge

Merge variants from multiple sources
Phenotype
Filter to gene lists based on ontology keywords
Population

Filter on population frequencies in public databases (gnomAD/Exac/1KG/UK10K) or number of samples in this database.

Tissue Expression

Filter based on tissue specific expression (from Human Protein Atlas)
Venn

A filter based on set intersections between parent nodes
Zygosity

Compound HET and other Zygosity filters


















