Sequencing Runs
Note: This feature may not be enabled on all systems as it requires access to a network drive (eg a diagnostic lab intranet)
VariantGrid can be setup to automatically scan network disks for sequencing runs to collect QC metrics, gene coverage and automatically load VCFs.
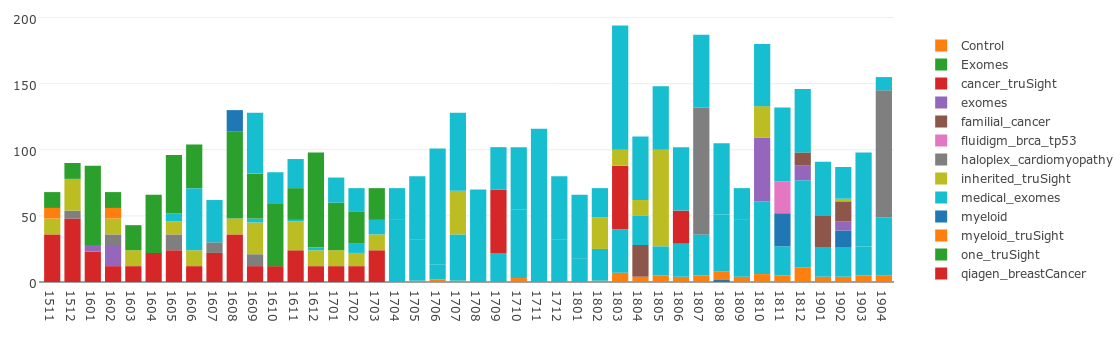 Sequencing Samples over time
Sequencing Samples over time
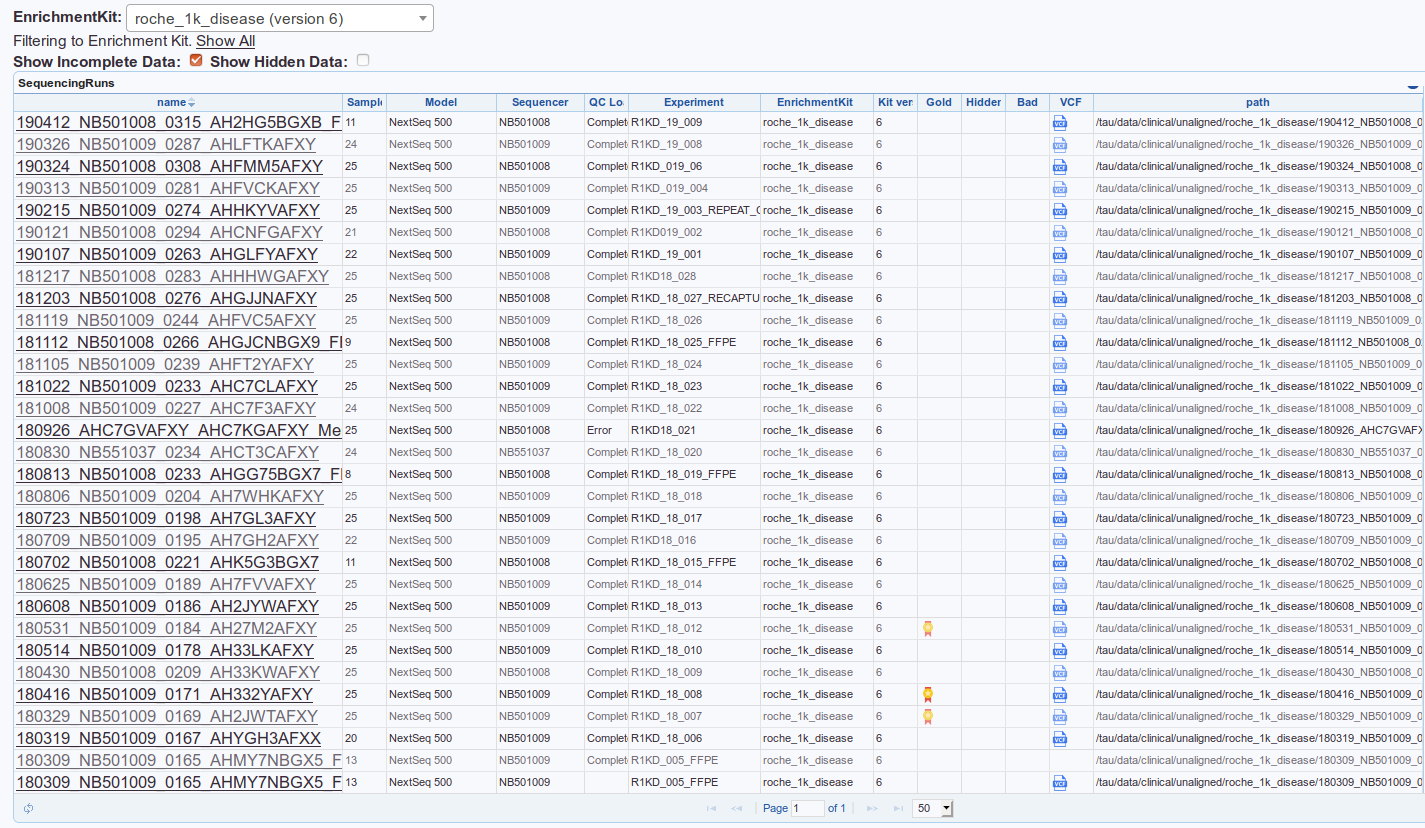 Automatically loaded sequencing runs + VCFs
Automatically loaded sequencing runs + VCFs
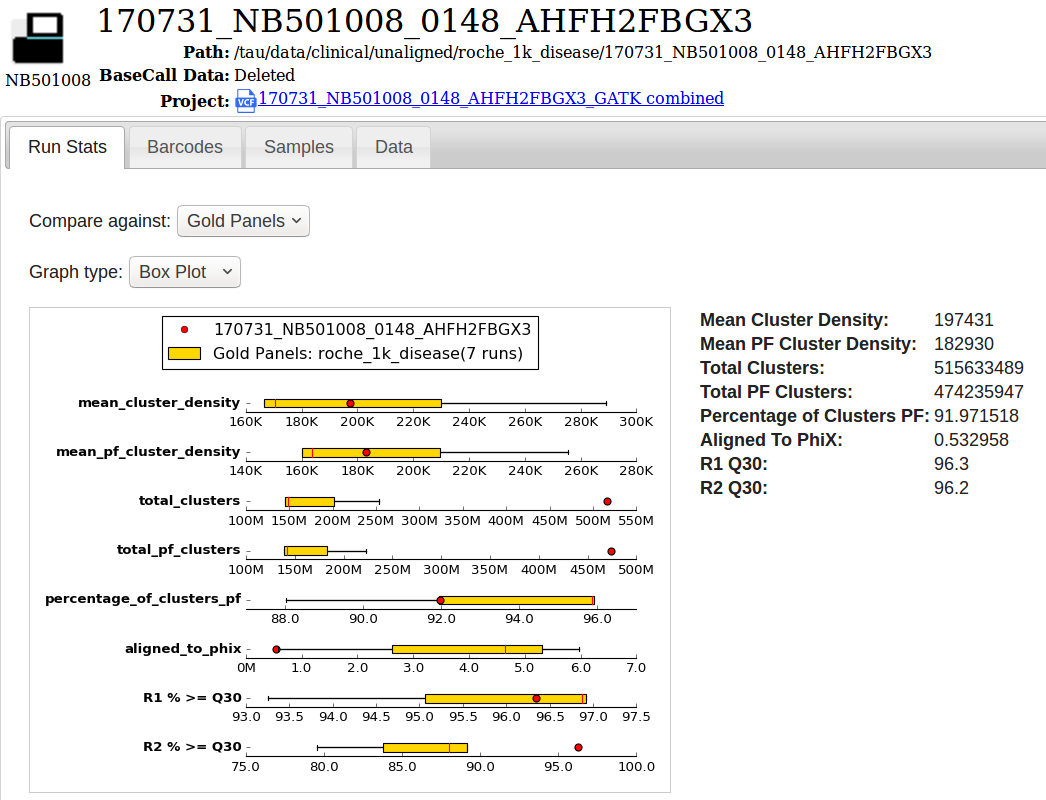 A Sequencing Run
A Sequencing Run
We collect Sequencing QC metrics and display them with interactive graphs. Collecting data over time allows us to see how this run compares to other runs over time (or vs gold standard runs).
Enrichment Kit
An EnrichmentKit is a lab method to enrich a sample for the DNA regions you are interested in. For instance an exome or custom gene capture kit, or amplicons.
You can set a bed file, a gene list and canonical transcript collection
See VariantGrid Admin docs for more information.
Gold Standard Runs
The administrator can mark a sequencing run as “Gold Standard” - which means it has passed validation / is of sufficient quality to be used as a benchmark for other runs.
Gold standard runs have an icon ( ) on the sequencing run grid.
) on the sequencing run grid.
Gold runs for an enrichment kit are used:
In boxplots on QC metrics pages for a sequencing run or other sample QC graphs.
To calculate average gene coverage on the GeneGrid page.
Finding sequencing data
Sequencing Runs are found by searching for the file ‘RTAComplete.txt’ on the server disks. You can ignore flow cells by putting a file “.variantgrid_skip_flowcell” in the directory.
Triggering a manual scan
Administrators, or users who have been give the permission “SeqAuto scan initiate” can
Menu: [sequencing], then manage disk scans link, then click the button Scan Disk for Sequencing Data